Hello there! 🙋🏼 I am Yizheng Wang (王一争), a Ph.D. candidate at the University of Electronic Science and Technology of China.
I obtained my bachelor’s degree 🎓 in Computer Science and Technology from the  School of Information Science and Engineering, Yanshan University (燕山大学,信息科学与工程学院).
School of Information Science and Engineering, Yanshan University (燕山大学,信息科学与工程学院).
I am pursuing my Ph.D. degree 🎓 through a combined Master’s and Doctoral program (硕博连读) at the  Institute of Fundamental and Frontier Sciences,
Institute of Fundamental and Frontier Sciences,  University of Electronic Science and Technology of China (电子科技大学,基础与前沿研究院), majoring in Computer Science and Technology.
University of Electronic Science and Technology of China (电子科技大学,基础与前沿研究院), majoring in Computer Science and Technology.
I am part of a joint training program (联合培养) at the  Yangtze Delta Region Institute (Quzhou),
Yangtze Delta Region Institute (Quzhou),  University of Electronic Science and Technology of China (电子科技大学,长三角研究院).
University of Electronic Science and Technology of China (电子科技大学,长三角研究院).
With the support of the China Scholarship Council, I am currently part of a joint training program (联合培养) at the School of Computer Science and Engineering,  University of New South Wales (新南威尔士大学,计算机科学与工程学院), conducting research at the Data and Knowledge Research Group.
University of New South Wales (新南威尔士大学,计算机科学与工程学院), conducting research at the Data and Knowledge Research Group.
I am currently conducting research at the Malab laboratory 🔬 under the supervision of Prof. Dr. Quan Zou (邹权教授) and Research Fellow Dr. Yijie Ding (丁漪杰研究员). I am responsible for a project funded by the  NSFC. My main research areas are machine learning 🤖 and computational biology 🧬, with several publications in SCI-indexed journals.
NSFC. My main research areas are machine learning 🤖 and computational biology 🧬, with several publications in SCI-indexed journals.
📒 Research Interests
🤖 Machine learning
- AI for science (科学智能)
- Large Language Model (大语言模型)
- Support Bio-sequence Machine (支持生物序列机)
- Support Bio-sequence Neural Network (支持生物序列神经网络)
🧬 Computational biology
- Biological sequence analysis (生物序列分析)
- Biological interactions prediction (生物关联预测)
- Multiple sequence alignment (多序列比对)
- Therapeutic peptide design (治疗肽设计)
- Phylogenetic tree construction (系统发生树构建)
🔥 News
- 2025.10.29: I have been awarded the 🎓 National Scholarship for Doctoral Students! 🎉
- 2025.09.17: My paper has been recognized by
 as a 🏆 Highly Cited Paper! 🎉
as a 🏆 Highly Cited Paper! 🎉 - 2025.09.11: I have received the official offer from
 and have enrolled as a joint training Ph.D. student! 🎉
and have enrolled as a joint training Ph.D. student! 🎉 - 2025.08.27: I have been funded by the
 NSFC Youth Science Fund Category C as Host, 300,000 CNY¥! 🎉
NSFC Youth Science Fund Category C as Host, 300,000 CNY¥! 🎉 - 2025.07.25: I attended 2025 International Conference on Intelligent Computing and gave an oral 🎤 presentation! 🎉
- 2025.05.13: My paper has been recognized by
 as both a 🏆 Highly Cited Paper and a 🔥 Hot Paper! 🎉
as both a 🏆 Highly Cited Paper and a 🔥 Hot Paper! 🎉 - 2024.12.27: I have been awarded the CSC Scholarship for the international ✈️ joint training program! 🎉
- 2024.10.07: My new paper has been accepted by the CCF A journal SCIENCE CHINA Information Sciences! 🎉
- 2024.03.17: My new personal homepage 🌐 has been released! 🎉
📝 Projects
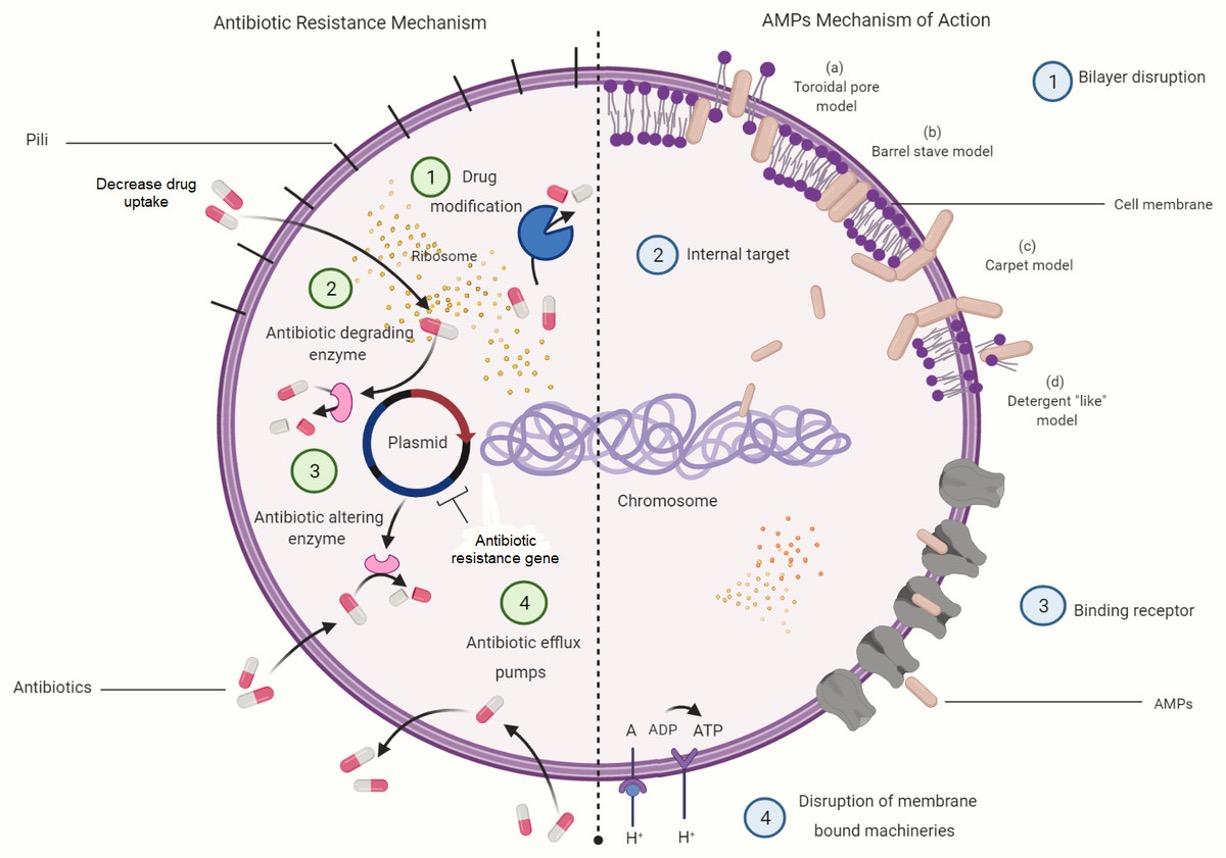
(1) Rational De Novo Design of Multifunctional Therapeutic Peptides Guided by Support Bio-sequence
Yizheng Wang, Yijie Ding*, Quan Zou*
Focuses on PDAC, aiming to develop multifunctional therapeutic peptides through rational de novo design guided by supportive bio-sequences.
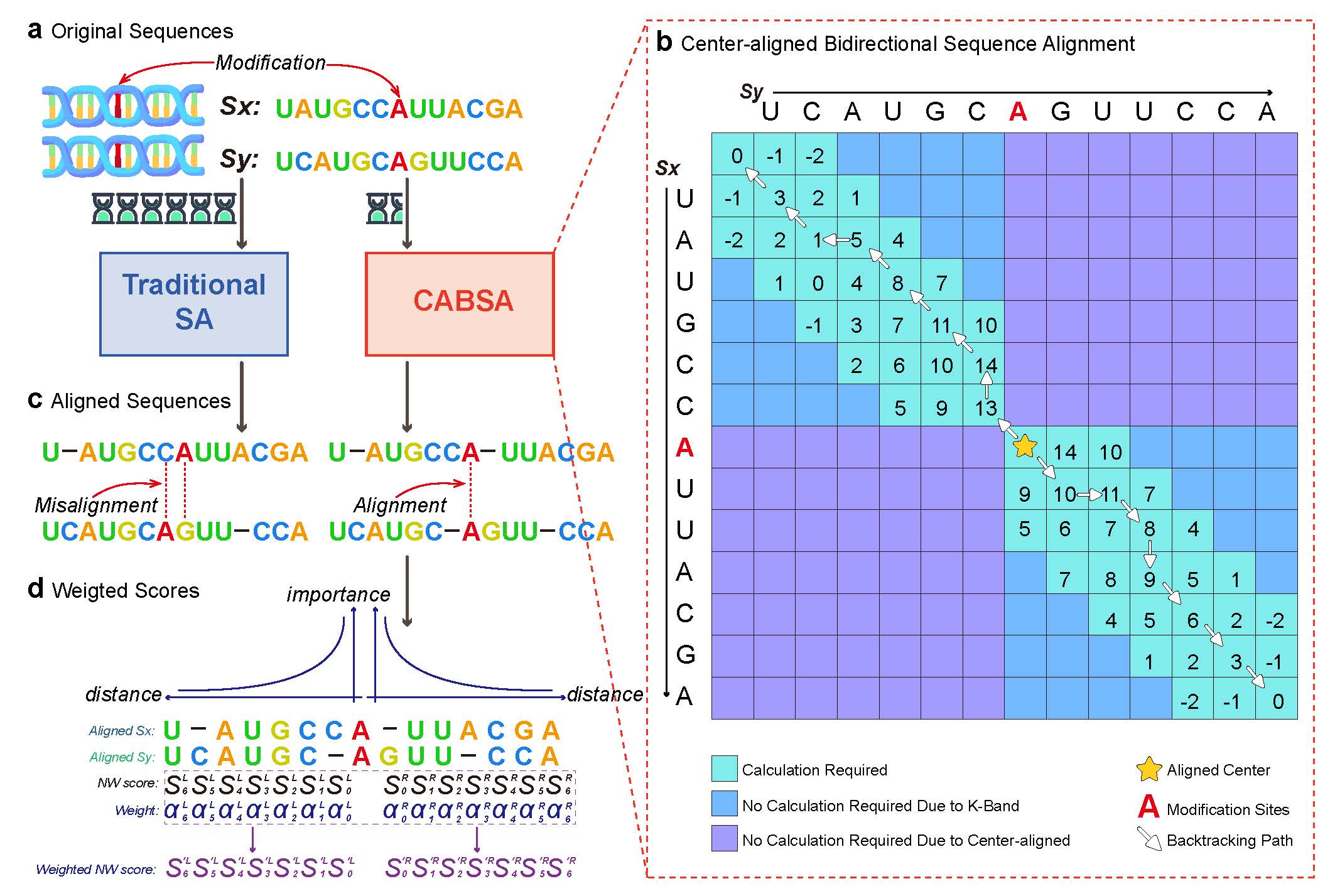
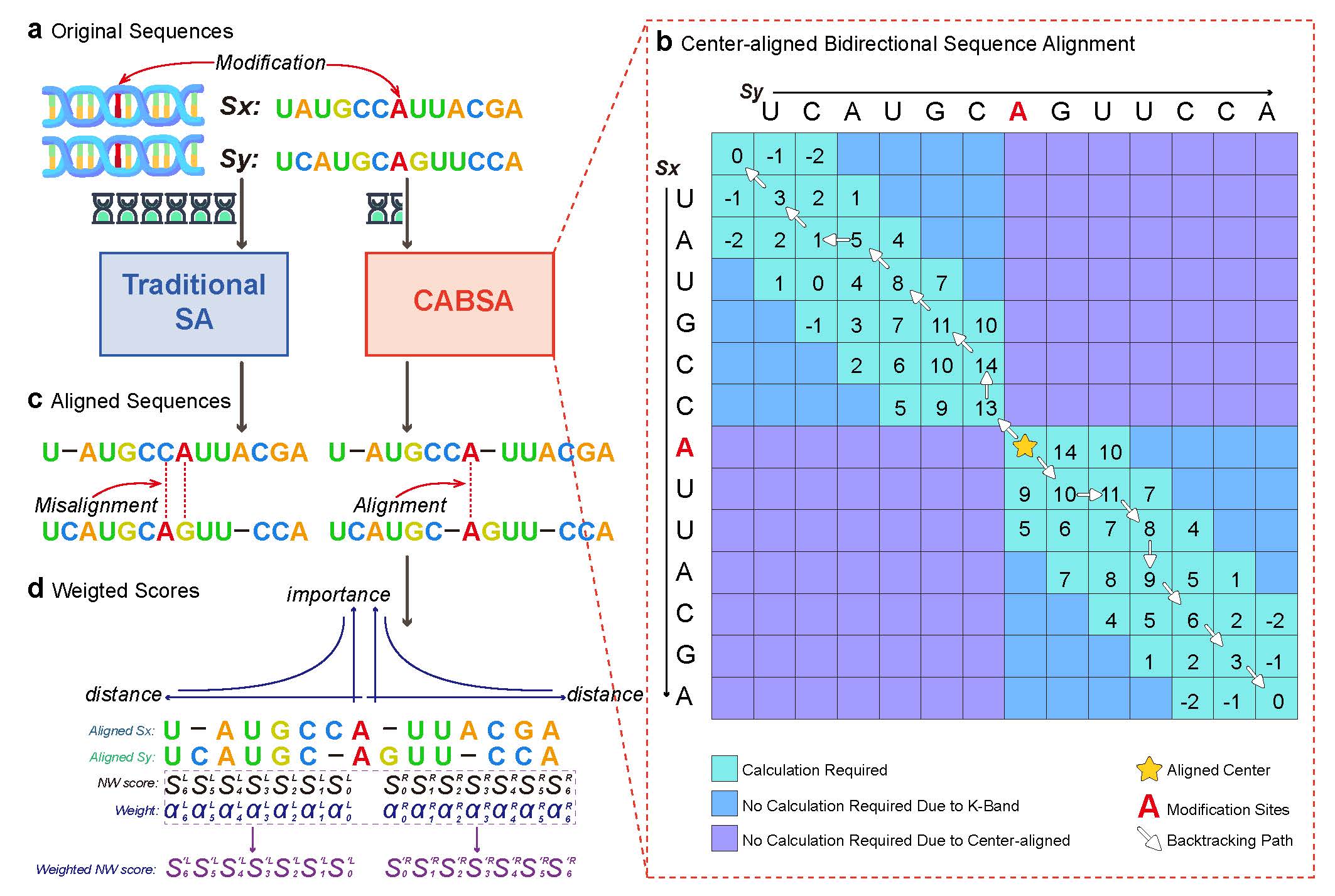
Yizheng Wang, Pinglu Zhang, Yi Ding, Wenyu Zhang, Yanming Wei, Yijie Ding*, Quan Zou*
SBNN advances the Support Bio-Sequence Machine (SBSM) framework toward bio-language modeling and epigenetic modification prediction.
Introduced a feature-free sequence learning approach with weighted multiple sequence alignment, enabling unified modeling of local and global dependencies in DNA, RNA, and protein sequences.
Evaluated on 76 datasets covering four modification types and 18 context lengths (11–500 nt), SBNN outperformed 17 state-of-the-art methods.
 Continuing Support Project of the Original Exploration Project of the NSFC
Continuing Support Project of the Original Exploration Project of the NSFC
Yizheng Wang, Yijie Ding*, Quan Zou*
A fast phylogenetic tree construction method based on a bio-large language model with siamese metric learning, trained on millions of phylogenetic trees.
Reformulated phylogenetic inference as a distance learning problem in representation space, achieving high accuracy with substantially improved efficiency for large-scale analyses.
* In ancient Chinese mythology, JianMu(建木) is a sacred tree, a bridge connecting heaven and earth. In biology, the phylogenetic tree is a bridge connecting genes and evolution. Thus, we chose the name JianMu for our model, highlighting its role in uncovering species relationships.
(4) SSM: Support Subsequence Machine
Yizheng Wang, Yijie Ding*, Quan Zou*
🌐 Website
Proposed a Support Bio-Subsequence Model that replaces global alignment with subsequence-level similarity, capturing functionally or structurally important local patterns beyond global sequence similarity.
Formulated support subsequence selection as a pattern mining problem, yielding an interpretable white-box model that quantifies subsequence importance and provides mechanistic biological insights.
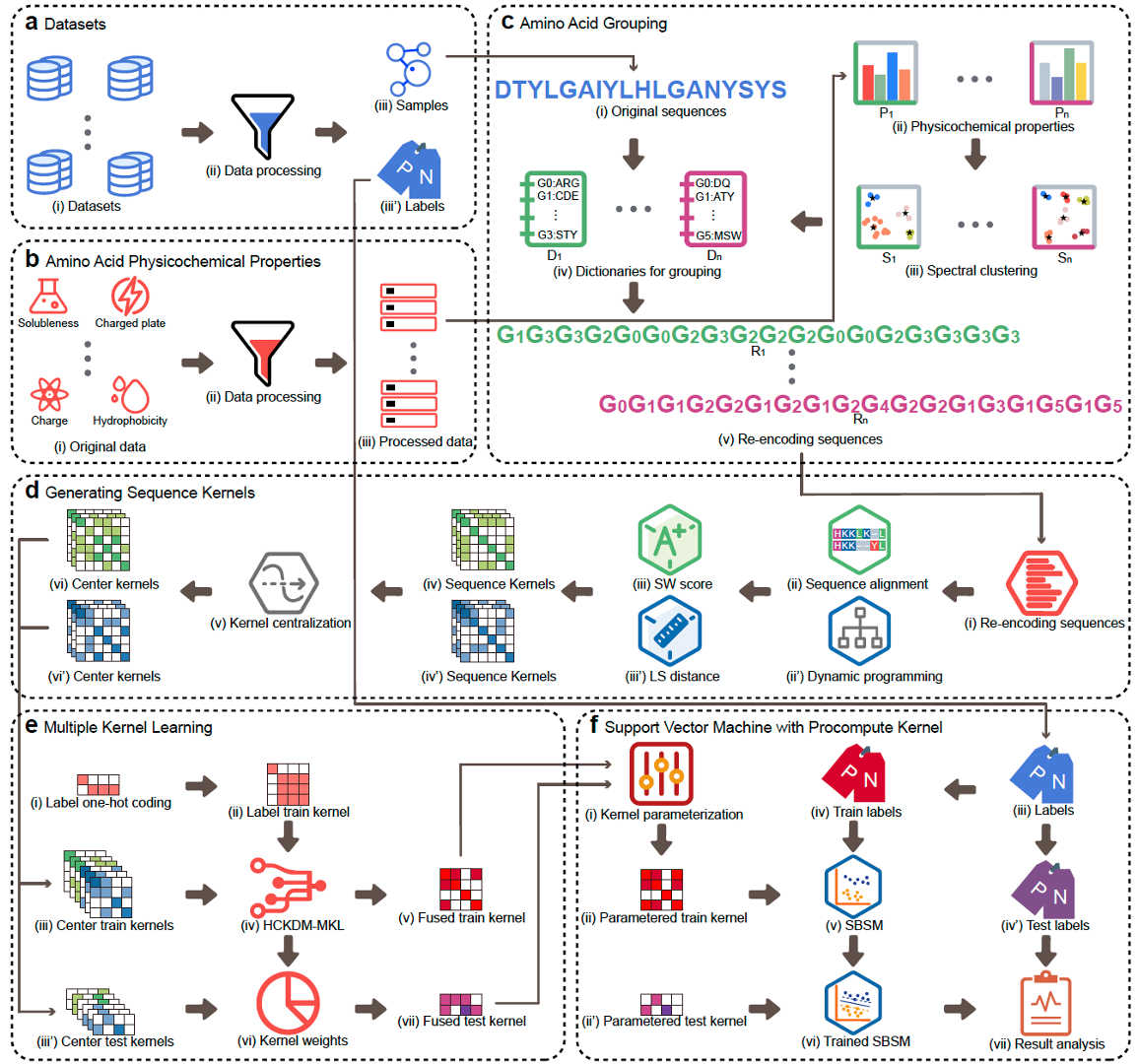
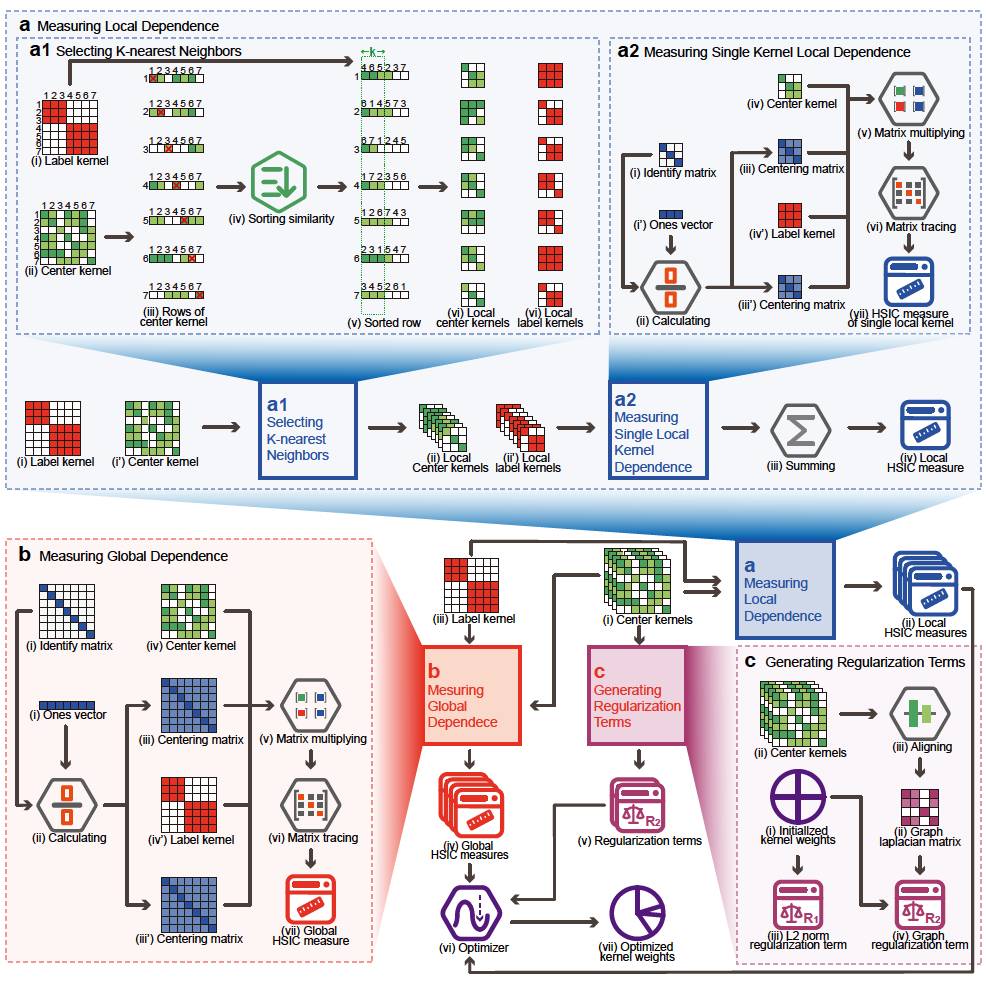
(5) SBSM-Pro: Support Bio-sequence Machine for Proteins
Yizheng Wang, Yixiao Zhai, Yijie Ding*, Quan Zou*
SCIENCE CHINA Information Sciences, CCF A, CAAI A, IF2024=7.300, 中科院1区top
 🏆 Highly Cited Paper (高被引论文) | 🔥 Hot Paper (热点论文)
🏆 Highly Cited Paper (高被引论文) | 🔥 Hot Paper (热点论文)
A novel framework that operates directly on raw protein sequences without explicit feature extraction, effectively addressing small-sample limitations of conventional machine learning and deep learning approaches.
Developed a PSD pipeline that introduces structural and functional priors at the sequence level, improving robustness and interpretability.
Introduced HCKDM-MKL, integrating global and local sequence kernels to achieve stable performance with strong generalization across protein function and modification tasks.
 Original Exploratory Project of the NSFC
Original Exploratory Project of the NSFC
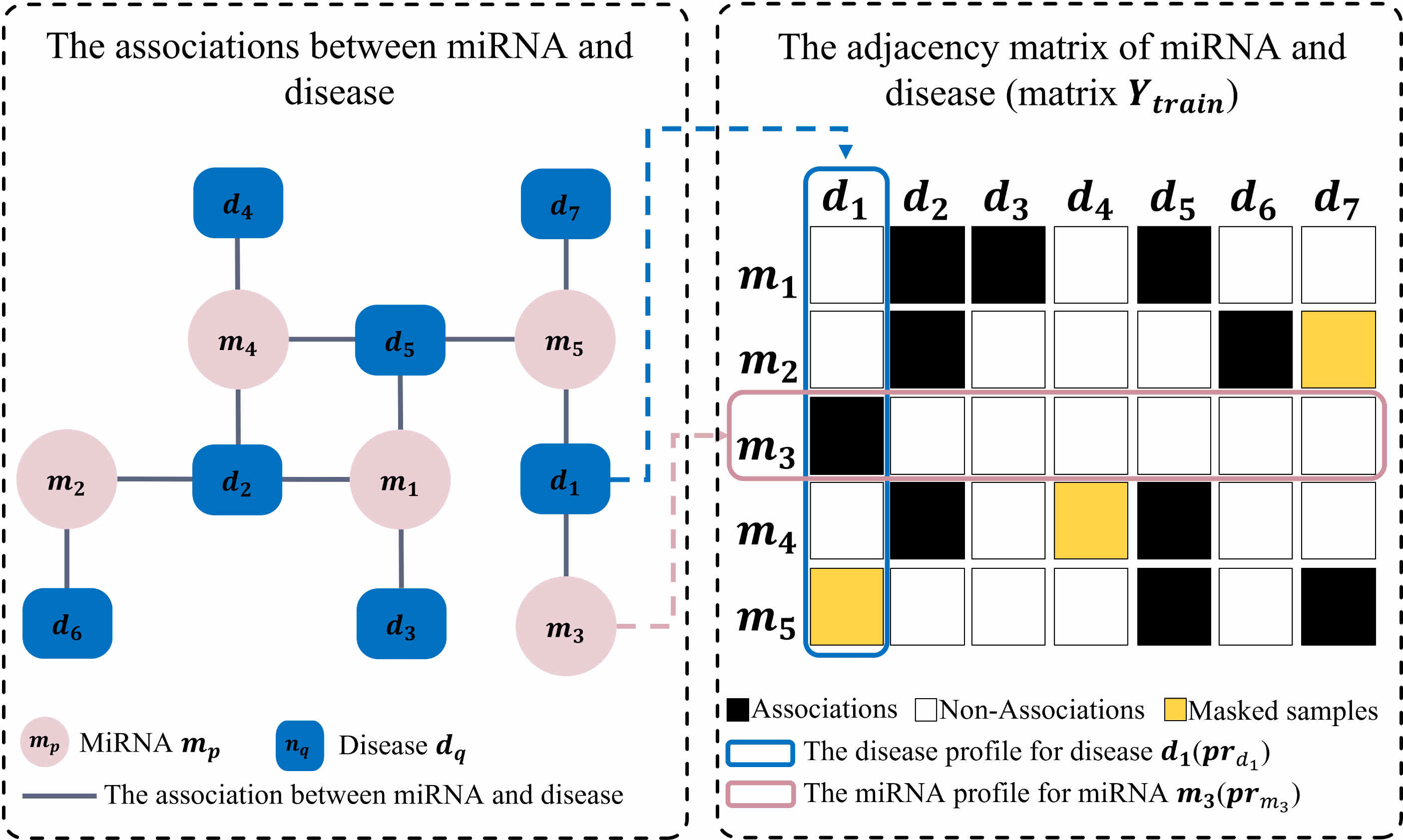
Yizheng Wang, Xin Zhang, Ying Ju, Qing Liu, Quan Zou, Yazhou Zhang, Yijie Ding*, Ying Zhang*
Frontiers of Computer Science, CCF B, CAAI B, IF2022=4.200, 中科院2区
 🏆 Highly Cited Paper (高被引论文)
🏆 Highly Cited Paper (高被引论文)
Proposed a low-rank approximation and multi-kernel learning–based link propagation method for efficient miRNA–disease association prediction, achieving high accuracy with reduced computational cost.
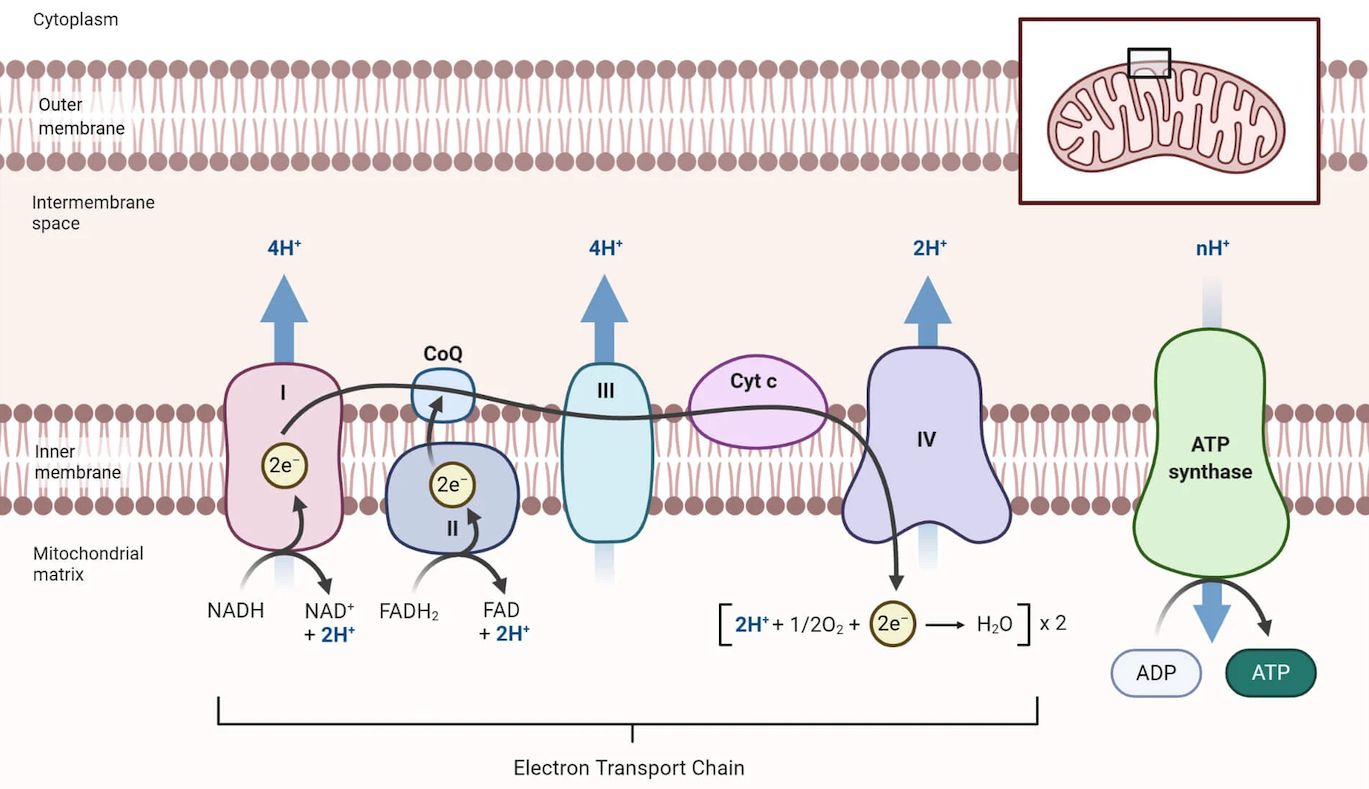
(7) ET-MSF: A model stacking framework to identify electron transport proteins
Yizheng Wang, Qingfeng Pan*, Xiaobin Liu*, Yijie Ding*
Frontiers in Bioscience-Landmark, JCRQ2, IF2021=4.009
Developed a computational approach to identify electron transport proteins, supporting studies of cellular respiration and mitochondrial function, and facilitating mechanistic research on diabetes, Parkinson’s disease, and Alzheimer’s disease.
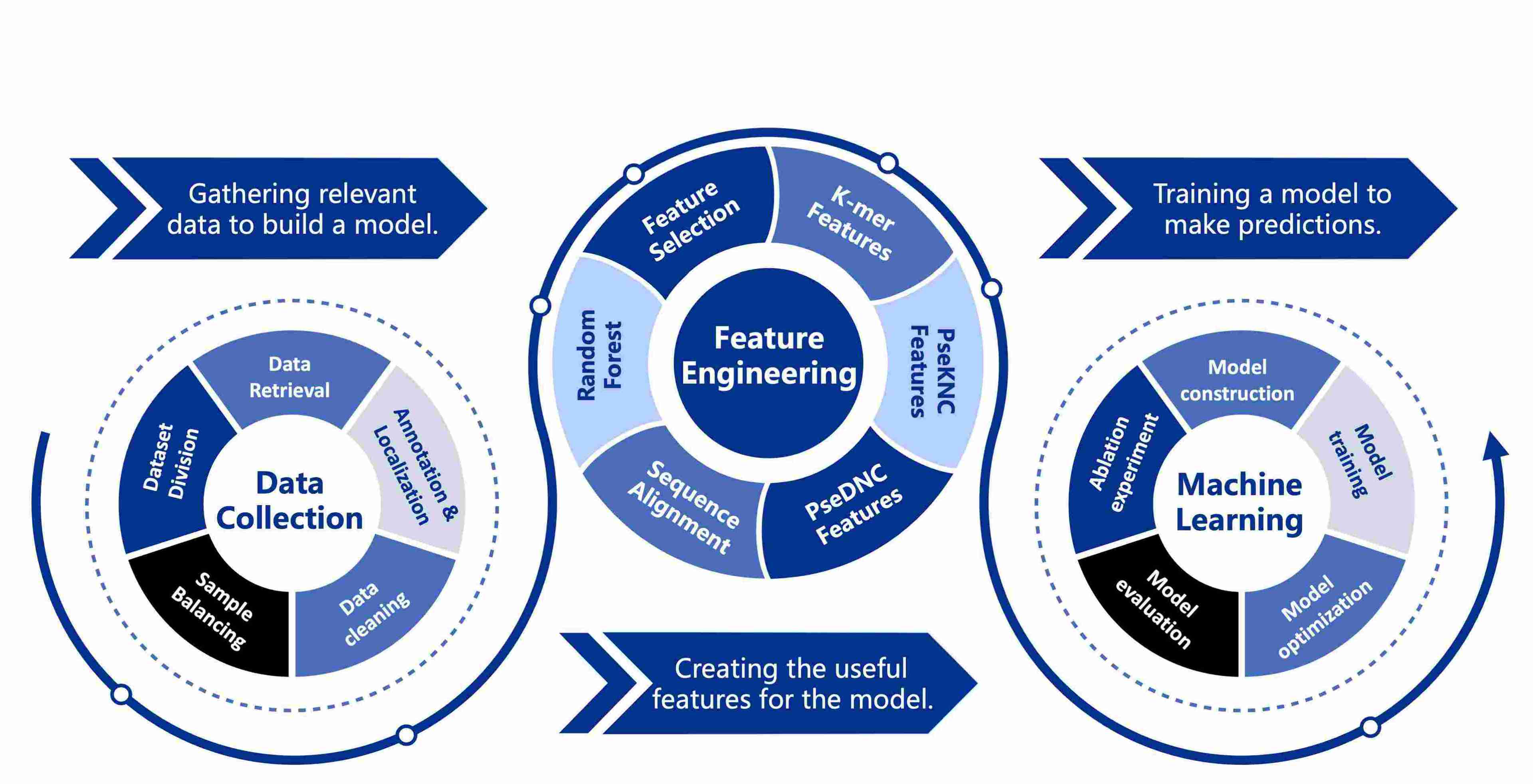
(8) SeqAlignXGBoost: Sequence Alignment and Feature Selection for m1A Modification Site Identification
Yizheng Wang, Yijie Ding*, Quan Zou*
2025 International Conference on Intelligent Computing (ICIC2025), CCF C
Developed a computational tool for identifying RNA N1-methyladenosine modification sites, supporting studies of RNA stability, translational regulation, and gene expression control, and facilitating epitranscriptomic research.
-
(9) A survey on multi-view fusion for predicting links in biomedical bipartite networks: Methods and applications, Yuqing Qian, Yizheng Wang, Junkai Liu, Quan Zou*, Yijie Ding*. Information Fusion, Q1, IF2024=14.700
-
(10) TPMA: A two pointers meta-alignment tool to ensemble different multiple nucleic acid sequence alignments, Yixiao Zhai, Jiannan Chao, Yizheng Wang, Pinglu Zhang, Furong Tang*, Quan Zou*. PLOS Computational Biology, Q1, IF2024=3.800
-
(11) DTI-RME: a robust and multi-kernel ensemble approach for drug-target interaction prediction, Yuqing Qian, Xin Zhang, Yizheng Wang, Quan Zou, Chen Cao, Yijie Ding*, Xiaoyi Guo*. BMC biology, Q1, IF2025=4.500
-
(12) HAlign-G: rapid and low-memory multiple-genome aligner for large-scale closely related genomes, Pinglu Zhang, …, Yizheng Wang, Quan Zou*, Furong Tang*, Ximei Luo*. Genome Biology, Q1, IF2025=9.400
-
(13) deMEM: a novel divide-and-conquer framework based on de Bruijn graph for scalable multiple sequence alignment, Yanming Wei, …, Yizheng Wang, Liang Yu*, Quan Zou*. GigaScience, Q1, IF2025=3.900
-
(14) Application of Artificial Intelligence in Drug-Target Interactions Prediction: A Review, Qian Liao, …, Yizheng Wang, Yijie Ding*, Quan Zou*, Ke Han*. npj Biomedical Innovations
📜 Patents
- 2025.04: A Phylogenetic Tree Construction Method Based on Metric Learning, (No.CN202510533729.3)
- 2024.09: A Method for Identifying Nucleic Acid Modification Sites Using Neural Network Models, (No.CN202410638063.3)
- 2023.11: A Construction Method for Biological Sequence Ensemble Classifiers and Methods for Predicting and Classifying Biological Sequences, (No.CN202310249336.0)
- 2023.03: A Training Method and Model for Predicting microRNA-Disease Associations, (No.CN202211444626.2)
🎖 Funds
- 2025.08: Research on Biological Sequence Analysis Combining Large Language Models and Metric Learning, Youth Science Fund Project, Category C (青年科学基金C类), Host,
 National Natural Science Foundation of China (国家自然科学基金委员会), 300,000 CNY¥
National Natural Science Foundation of China (国家自然科学基金委员会), 300,000 CNY¥ - 2021.11: Intelligent Question-Answering System for COVID-19 Medical Knowledge, The National College Student Innovation and Entrepreneurship Training Program (国家级大学生创新创业训练计划项目), Ministry of Education of the P.R.C. (教育部), 30,000 CNY¥
🏅 Awards
- 2025.10: National Scholarship for Doctoral Students (博士研究生国家奖学金), Ministry of Education of the P.R.C. (教育部), 30,000 CNY¥
- 2025.10: First-Class Academic Scholarship (一等学业奖学金), University of Electronic Science and Technology of China, 12,000 CNY¥
- 2025.09: Tuition Fee Scholarship (学费奖学金),
 University of New South Wales, Sydney (新南威尔士大学), Full tuition waiver
University of New South Wales, Sydney (新南威尔士大学), Full tuition waiver - 2024.12: CSC Scholarship for Study Abroad (公派留学奖学金), China Scholarship Council (国家留学基金管理委员会), 30,000 AUD$
- 2022.06: Outstanding graduates of Hebei Province (河北省优秀毕业生), Hebei Provincial Department of Education (河北省教育厅)
📖 Educations
- 2024.09 - (now): Ph.D. candidate, Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China (电子科技大学,基础与前沿研究院)

- 2025.11 - (now): Joint training student, School of Computer Science and Engineering, University of New South Wales (新南威尔士大学,计算机科学与工程学院)

- 2023.07 - (now): Joint training student, Yangtze Delta Region Institute (Quzhou), University of Electronic Science and Technology of China (电子科技大学,长三角研究院)

- 2022.09 - 2024.06: Master, Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China (电子科技大学,基础与前沿研究院)

- 2018.09 - 2022.06: Bachelor, School of Information Science and Engineering, Yanshan University (燕山大学,信息科学与工程学院)

- 2015.09 - 2018.06: High school student, Hebei Zhengding Senior High School, East Campus (河北正定中学,东校区)

